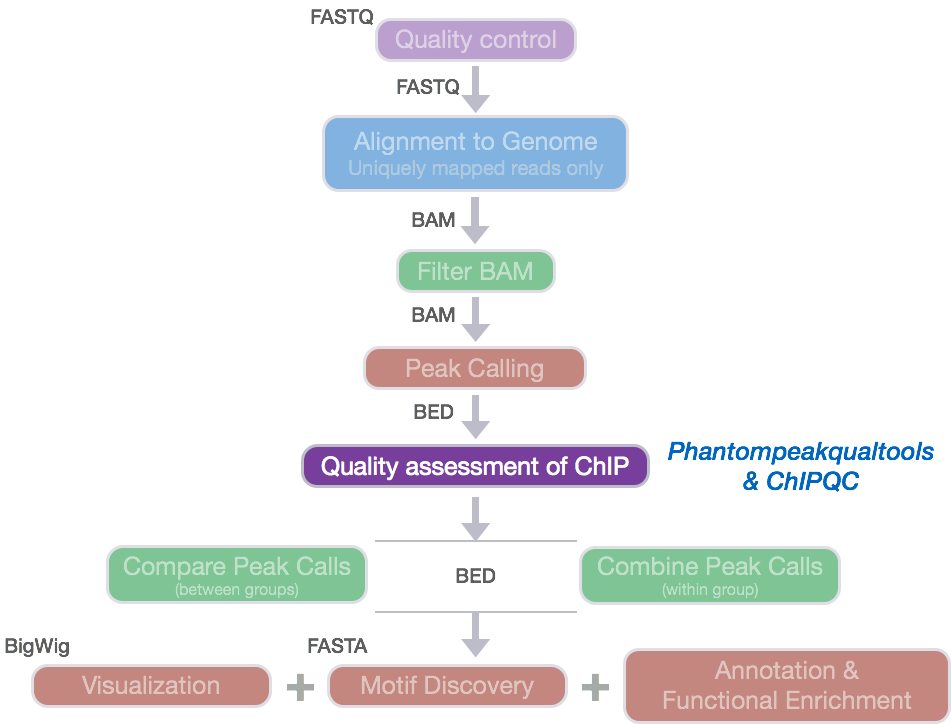
TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance
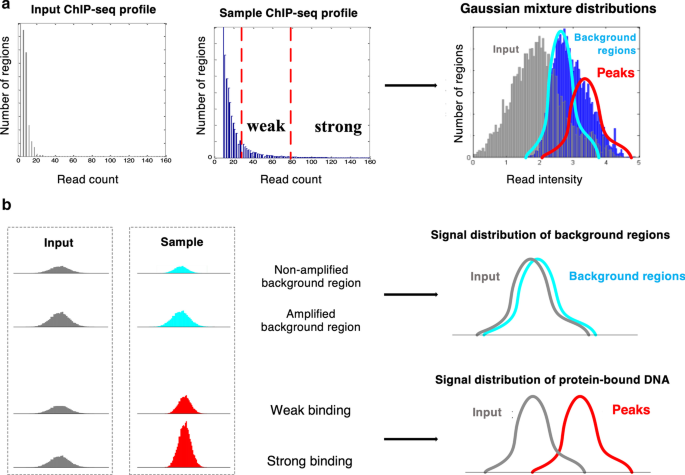
ChIP-BIT2: a software tool to detect weak binding events using a Bayesian integration approach | BMC Bioinformatics | Full Text
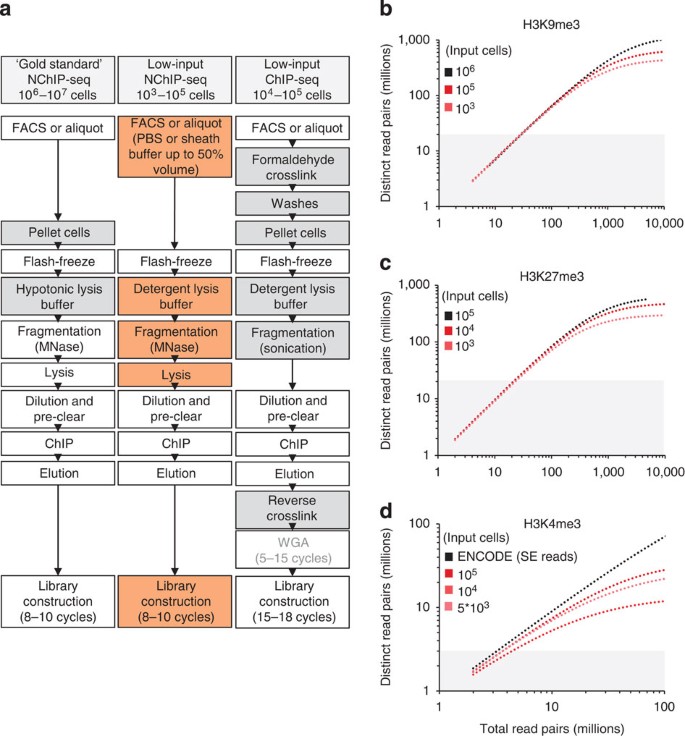
An ultra-low-input native ChIP-seq protocol for genome-wide profiling of rare cell populations | Nature Communications
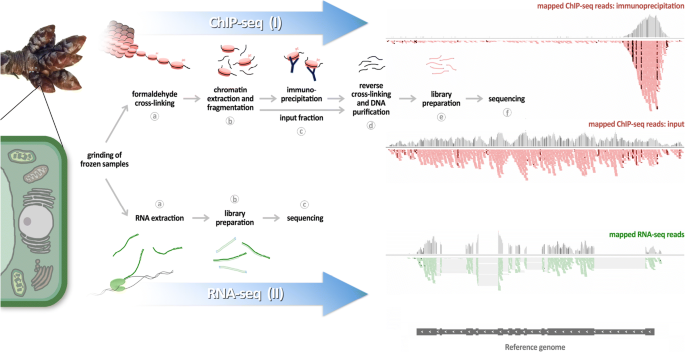
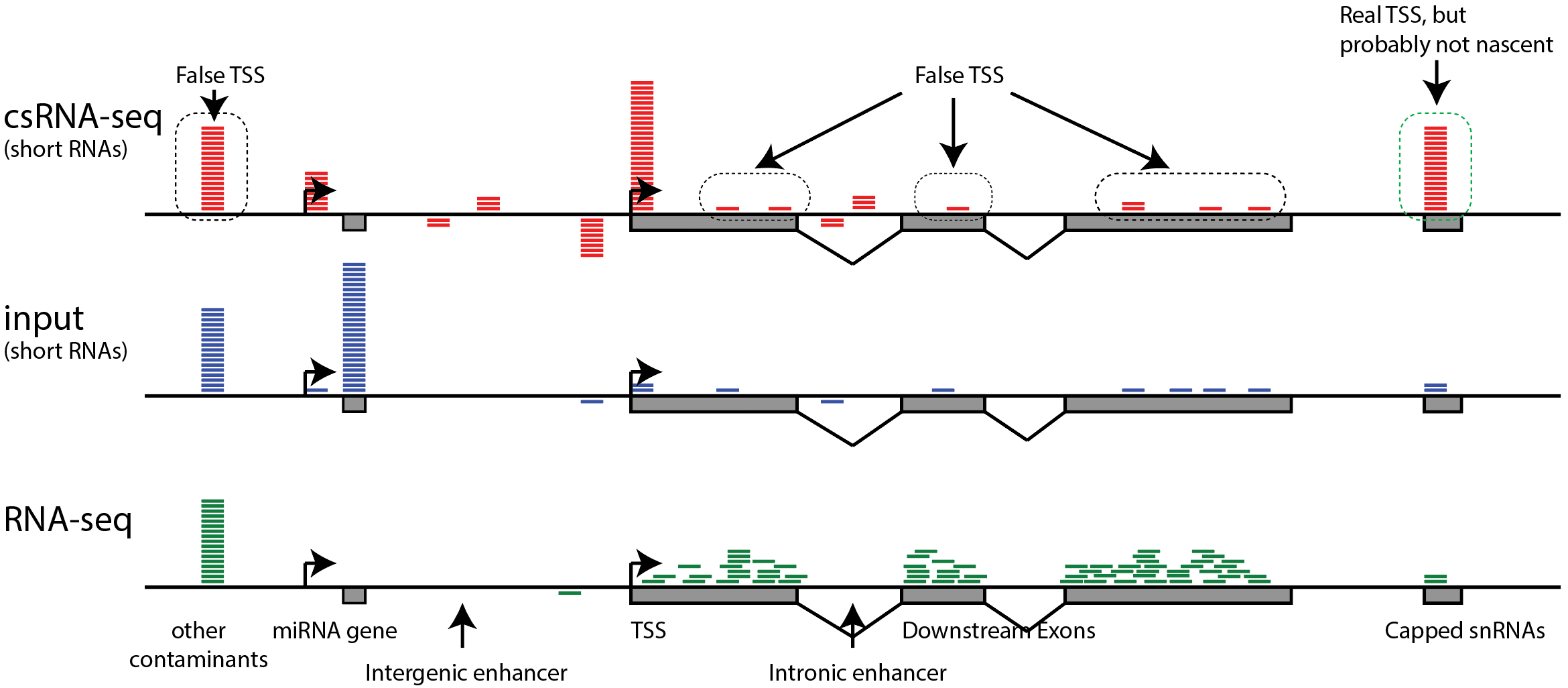
ChIP-seq and RNA-seq for complex and low-abundance tree buds reveal chromatin and expression co-dynamics during sweet cherry bud dormancy | SpringerLink
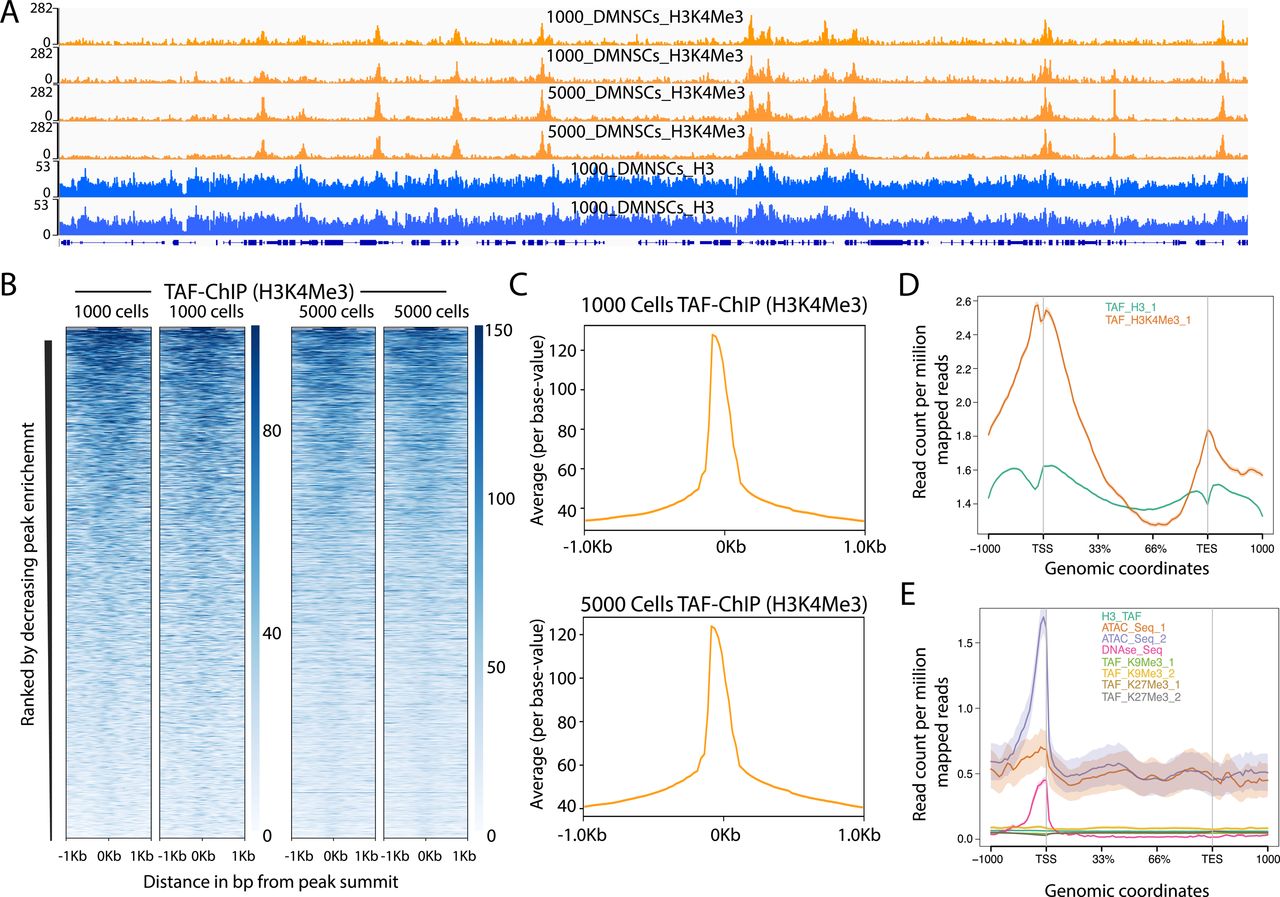
TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance

TAF-ChIP: an ultra-low input approach for genome-wide chromatin immunoprecipitation assay | Life Science Alliance

ChIP-seq protocol for sperm cells and embryos to assess environmental impacts and epigenetic inheritance - ScienceDirect
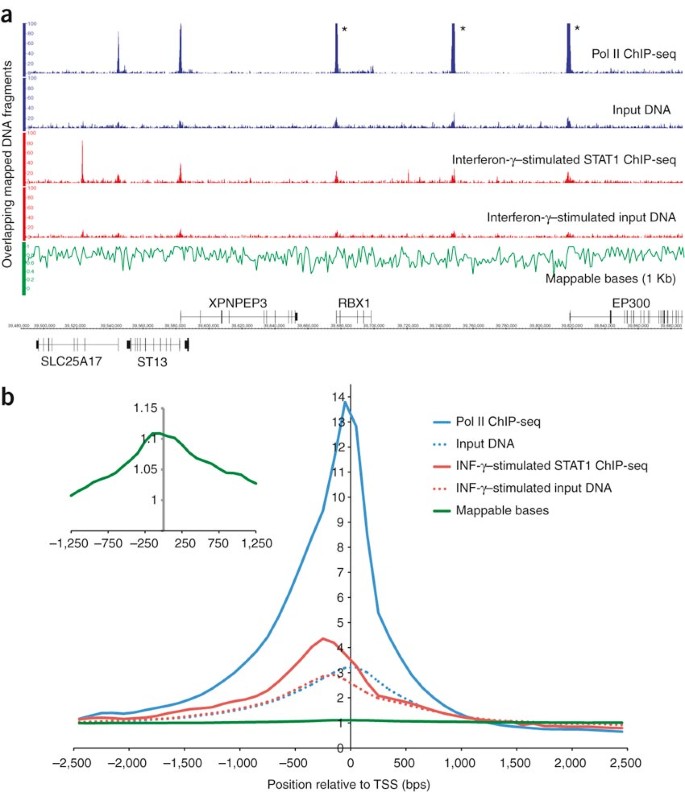
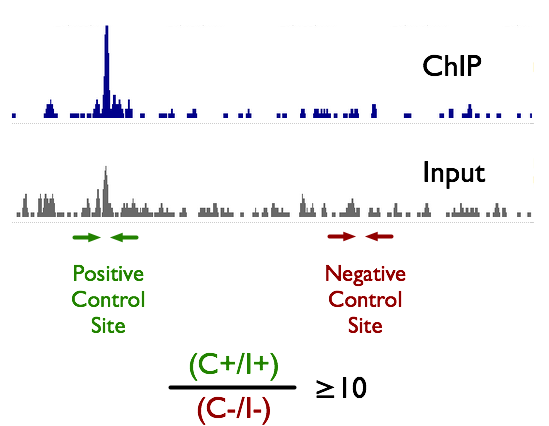
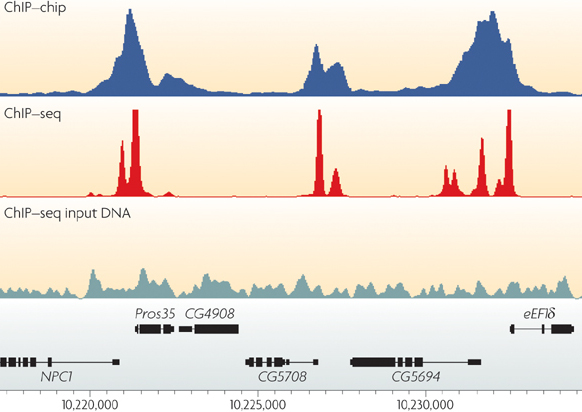
PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls | Nature Biotechnology